

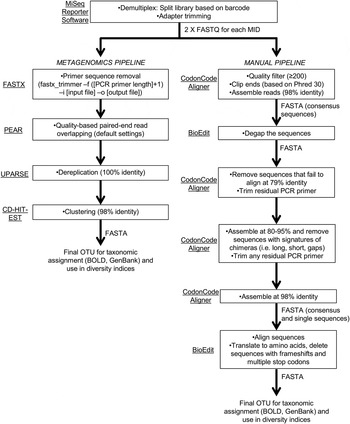
Parts of fragments containing repetitive DNA are therefore commonly lost during cloning 11. Moreover, the superhelical stress induced by the presence of repeats generates secondary structures that become substrates for deletion. Fragments rich in repeats affect the structure of DNA, forming hairpins and other complex structures that induce instability 10. Another problem is the extreme difficulty in cloning highly repetitive DNA into plasmid vectors 9. New bioinformatic strategies are under development to solve these problems 8 but so far these have found the assembly of long stretches beyond their capacity. In addition, numerous copies of reads can be nearly identical, leading to a tendency to assemble them into single, collapsed contigs. The sequence reads obtained with NGS platforms are short and simply do not span long repetitive sequences. Indeed, even though next-generation sequencing (NGS) has provided vast amounts of genomic sequence data, many genomes are not fully known because of the difficulty in assembling repeated elements 7. melanogaster, the genome of which is commonly regarded as completely sequenced 5.įailure to assemble long stretches of repeats accounts for most of the gaps that remain in “sequenced genomes”, including the human genome 6. These repetitive regions are poorly analyzed even in model organisms such as D. However, fluorescent in situ hybridisation (FISH) experiments have shown that some plant SSRs loci are organized into large clusters, usually associated with the constitutive heterochromatin 4. Most studies have relied on the analysis of SSRs located in coding regions of euchromatin in which the size of the repeats sequenced is around 100 bp. These sequences have also aroused attention given their apparent involvement in the onset of some heritable human disorders 3. Given their high degree of polymorphism, SSRs have been widely used as molecular markers in mapping, DNA fingerprinting and genetic evolution studies. Microsatellites, or simple sequence repeats (SSRs), are a type of repeat made up of units of up to 6 bp arranged in tandem arrays of very variable size distributed throughout the genome 2. There are three main classes of repeat: transposable elements, tandem repeats, and high copy number genes (such as ribosomal genes). Repetitive DNA is present in most organisms in variable proportion in some it is actually the most abundant component of genomic DNA 1. To demonstrate that this procedure can sequence other kinds of repetitive DNA, it was used to examine a 4.5 kb fragment containing a cluster of 15 repeats of the 5S rRNA gene of barley. This strategy allowed the sequencing of a fragment containing a stretch of 6.2 kb of continuous SSRs. This paper reports an alternative method for sequencing of long stretches of repetitive DNA based on the combined use of 1) a linear vector to stabilize the cloning process, and 2) the use of exonuclease III for obtaining progressive deletions of SSR-rich fragments. Indeed, most sequenced genomes contain gaps owing to repetitive DNA-related assembly difficulties. Unfortunately, the molecular characterization of these repeats has been hampered by technical limitations related to cloning and sequencing. Variation in repeats can alter the expression of genes, and changes in the number of repeats have been linked to certain human diseases. In some species, SSRs may be organized into long stretches, usually associated with the constitutive heterochromatin. SSRs are a type of repetitive DNA formed by short motifs repeated in tandem arrays. Repetitive DNA is widespread in eukaryotic genomes, in some cases making up more than 80% of the total.


 0 kommentar(er)
0 kommentar(er)
